Much of the activities in our invertebrate collections are dedicated to so-called DNA-barcoding. Our mission in the NORBOL consortium is to produce DNA-barcodes, particularly for marine fauna in Norwegian waters and to make these barcodes available with open access to records and metadata in Boldsystems.com. In the same manner we have also worked to produce DNA-barcodes for marine invertebrates on the West-African continental shelf in a project called we call MIWA.
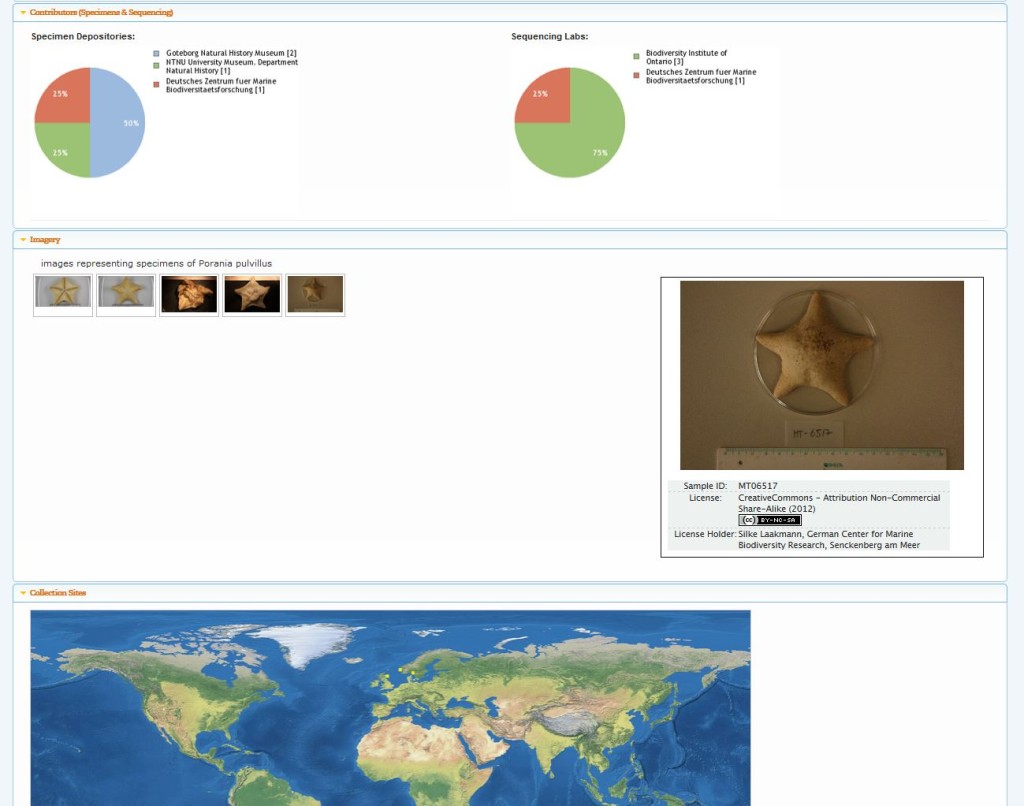
These QR-codes will take you to maps with plots of specimens that have been barcoded in our projects (or simply click on them. The red dots on the maps are interactive):
The basic idea motivating these activities is very simple in principle. You collect specimens and identify them, preferably to species, take digital photographs, and upload information about collection site and other relevant data to a database (BOLDsystems.org).
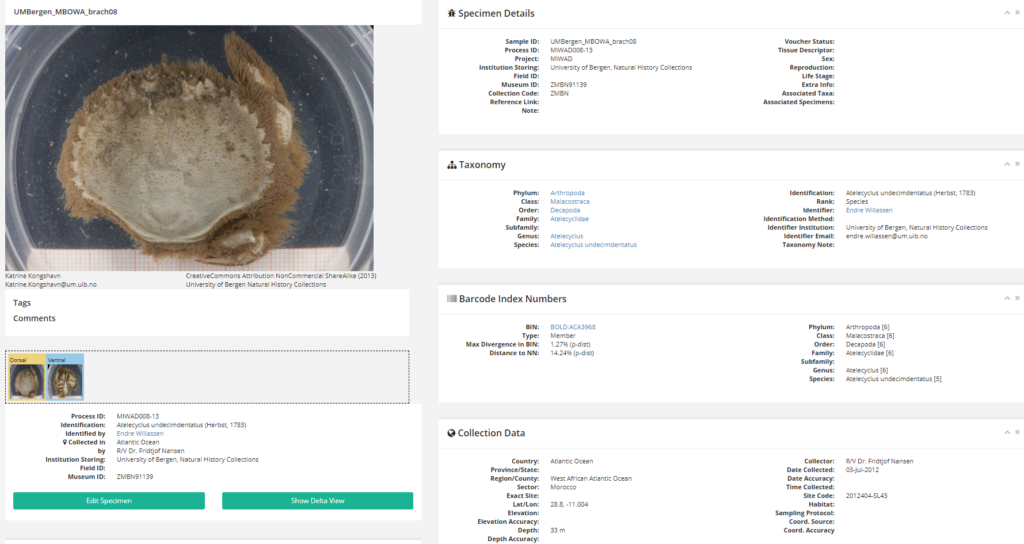
The specimen page has a picture and other data about the organism that the DNA sequence (presumably) was produced from (click picture to enlarge).
You take a tissue sample to extract DNA from the specimen and use DNA-sequencing technology to target a special fragment of DNA to read the sequence of nucleotides. The expectation is that this sequence may be unique for the particular species you identified. If indeed the expectation is fulfilled, you can use that sequence as an unambiguous identifier (“bar-code”) of that species. You have produced a DNA-barcode!
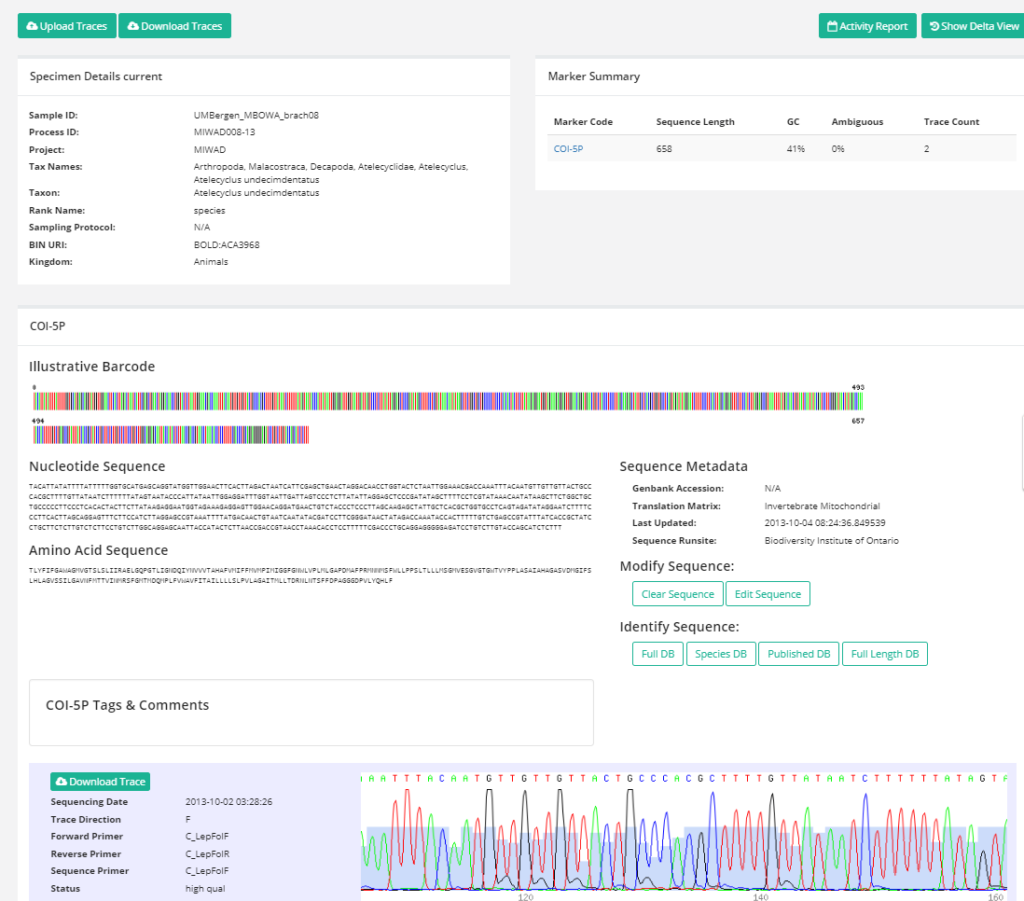
A sequence page in BOLD contains the DNA-sequence (the barcode), the aminoacid translation of the sequence, and the trace-files from the DNA-sequencing machine.(Click picture to enlarge)
Your barcode should enter a DNA-barcode library so that, with an appropriate web-interface to a powerful computer with a search algorithm that compares similarities, you should be able to search with a second sequence from another individual of the same species and find that it is identical, or at least very similar to the one you produced for the DNA-barcode library. The benefits are potentially many. One advantage is that you may be able to identify a species although all the morphological characteristics have been lost. For the biologist DNA-barcodes may help to identify juvenile stages of a species or even the stomach contents of a predator or scavenger. For conservation, customs, trade, and food authorities DNA barcodes are a powerful means to monitor resource exploitation and attempts to swindle with species identities or area of origin of biological products.
DNA-barcoding certainly also contributes to the mapping of species distributions and to survey genetic characteristics of taxa. Perhaps initially somewhat unexpected, it also reveals many problems in taxonomy that call for resolution through closer studies. More about this will follow in other blog posts.
-Endre